Aquaporins (AQP), channel proteins for water and glycerol, have been shown to play a role in maintaining cellular homeostasis, metabolism and multiple pathologies. Research by the Medical Structural Biology group at Lund University, led by Karin Lindkvist-Petersson, used various techniques to determine the structure of human aquaporin 7 (AQP7) and identify an inhibitor with potential therapeutic properties.
A common feature in biology is the structure-function relationship; how the shape of proteins are directly related to how they work and function. In Sweden, there are multiple large-scale infrastructures such as MAX IV, ESS and SciLifeLab that both national and international researchers from academia, industry and healthcare can access. One area of research that utilizes all three is that of Structural Biology which looks at this structure-function relationship on the molecular level.
Determining a 3D structure of AQP7 using X-rays
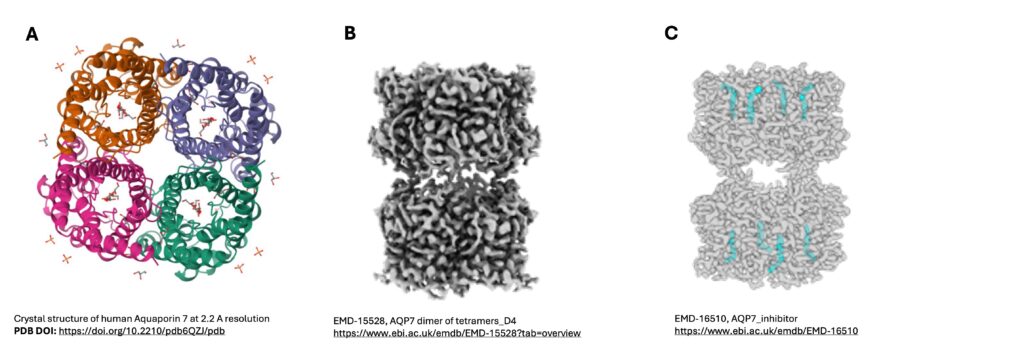
The aquaglyceroporin AQP7 is highly expressed in adipocytes, whereby it enables the efflux of glycerol and water, but a few years ago its role in transporting water was still debated. In order to address this question, the researchers wanted to determine the three-dimensional (3D) structure of AQP7 (deMaré at al., 2020). Macromolecular crystallography (MX) is a technique whereby the 3D structure of a protein is determined by exposing crystals of the protein of interest to X-rays. Screening of crystals was conducted at the BioMAX beamline, MAX IV, while data collection was done at PETRA III, DESY. Now that BioMAX is well established, the research group has done subsequent research at MAX IV. Structure determination and subsequent molecular dynamic simulations demonstrated that AQP7 (Figure 1A) has both glycerol and water molecules lining its pore, but the presence of glycerol prevents unrestricted transport of water across the membrane, as well as providing additional details about glycerol efflux.
Single particle cryo-EM structure of AQP7 reveals an additional role as junction protein
Following on from this, the researchers identified that human AQP7 is present in the pancreas with decreased expression in Type 2 Diabetes (Huang et al., 2023). In order to study AQP7 further, cryo-electron microscopy (cryo-EM) was utilized. Cryo-EM is an imaging technique to determine the 3D structure of molecules at cryo temperatures, which does not require crystallization of the protein. At the Cryo-EM unit, SciLifeLab Stockholm, a 2.55Å single particle cryo-EM structure of AQP7 was obtained (Figure 1B). Identification of extracellular loops connecting the two tetramers of AQP7 provided a molecular basis of how AQP7 interacts with other cells and molecules and that it acts as a junction protein. Cryo-EM also allowed for analysis of the central pore, which was not possible in the X-ray structure, due to its 4-fold symmetry.
Combined 3D structure of AQP7 and an inhibitor using cryo-EM
In addition to roles in cellular regulation and diabetes, reduced expression of AQP7 has been linked to improved prognosis in cancer but no therapeutics are available to date. Returning to the cryo-EM unit in SciLifeLab, the researchers produced a 3D structure of human AQP7 but now in complex with the inhibitor Z433927330 (Huang et al., 2024). This time the 3D structure allowed them to visualize where and how the inhibitor and protein interacted (Figure 1C), especially when compared to the previously determined 3D structure of AQP7 without inhibitor. In order to investigate the selectively of the inhibitor with AQP7, MD simulations and a computational analysis tool, AlphaFold2, was used to model other AQPs with the inhibitor. With the combined X-ray and cryo-EM structures of AQP7 and Z433927330, this opens the way for structure-based drug design to develop novel therapeutics.
Sweden facilitates structural biology research through large scale research infrastructures, dedicated platforms for proteins and expert personnel.
The cryo-EM unit at SciLifeLab is part of the Integrated Structural Biology platform which also includes the Swedish NMR Centre and Structural Proteomics, allowing for structure determination, protein interaction analysis and conformational dynamics using nuclear magnetic resonance and mass spectrometry respectively. In addition to the BioMAX beamline at MAX IV, the newly opened MicroMAX beamline allows for serial crystallography and the possibility to follow proteins in time. Along the lines of structure-based drug design, the X-ray crystallographic fragment screening platform, FragMAX, is available at MAX IV. When ESS opens for users, it will be possible to conduct macromolecular crystallography using neutrons instead of X-rays at the NMX instrument. If researchers are doing neutron experiments in other facilities before this, the Deuteration and Macromolecular Crystallisation (DEMAX) lab is open and provides services for providing deuterated molecules and support for protein crystallography.
For both MX and cryo-EM experiments, sample preparation is important as is the post data analysis which can involve multiple tools. Oftentimes, the research group themselves will prepare the samples but there are dedicated platforms which assist in protein production, crystallization and data analysis. These include the Protein Production Sweden infrastructure with sites all over Sweden and the National Bioinformatics Infrastructure (NBIS) which has allocated staff for cryo-EM. SciLifeLab regularly hosts events and training courses focused on cryo-EM, which can be found on the events calendar. InfraLife hosted a 2-week course on Integrated Structural Biology which covered 7 techniques, 7 site visits and 3 locations in Sweden. One of the authors in the case described here was an attendee in 2022. The ISB platform will now host the course from 2024 onwards.
Further information
Contact information
If you like to know more about how SciLifeLab, MAX IV and ESS can help with your Life Science needs, reach out to the Project Coordinators (Claire Lyons, claire.lyons@maxiv.lu.se or Josefin Lundgren Gawell, josefin.lundgren.gawell@scilifelab.se).
For more information about the aquaporin 7 research, please read the featured publications below or contact Karin Lindkvist-Petterson (karin.lindkvist@med.lu.se).
References
Sofia W. de Maré, Raminta Venskutonytė, Sandra Eltschkner, Bert L. de Groot, Karin Lindkvist-Petersson. Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7, Structure, Volume 28, Issue 2, 2020, Pages 215-222.e3, ISSN 0969-2126, https://doi.org/10.1016/j.str.2019.11.011. https://www.rcsb.org/structure/6QZJ
Huang, P., Venskutonytė, R., Prasad, R.B. et al. Cryo-EM structure supports a role of AQP7 as a junction protein. Nat Commun 14, 600 (2023). https://doi.org/10.1038/s41467-023-36272-y. https://www.ebi.ac.uk/emdb/EMD-15528?tab=overview .
Peng Huang, Hannah Åbacka, Carter K. Wilson, Malene Lykke Wind, Michael Rutzler, Anna Hagström-Andeersson, Pontus Gourdon, Bert L. de Groot, Raminta Venskutonyté, and Karin Lindkvist-Petersson. Molecular basis for human aquaporin inhibition. PNAS, 2024, Vol. 121, No. 7, e231968212. https://doi.org/10.1073/pnas.2319682121. https://www.ebi.ac.uk/emdb/EMD-16510
Are you interested in using a research infrastructure, but not sure where to go or who to talk to?
